|
|||
|
|||
This website is a tutorial for my informal Python beginners' course. We will use materials of DNA analyses as follows:
I believe writing and running Python programs are more important than learning Python syntaxes. We do not need to remember all syntaxes. In this website, the following points are my recommendations:
|
|||
|
|||
| Installing Python We will install python3. |
|||
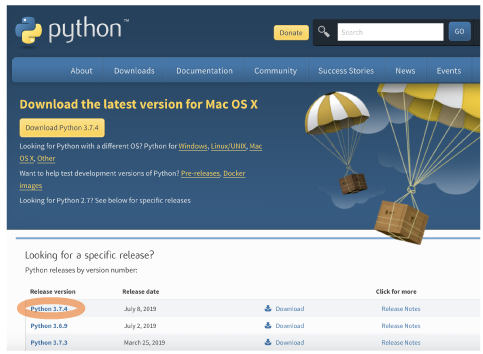
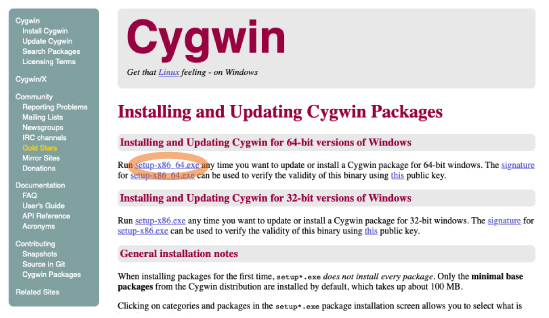
| [Mac, Win] Open the following website and click the latest link in the "Release version list": https://www.python.org/downloads/ |
|||
 |
|||
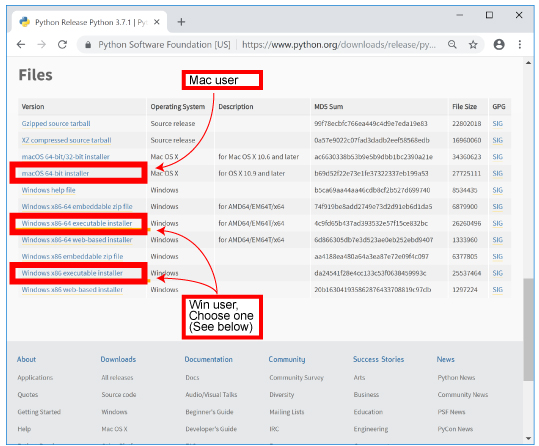
| Scroll down and choose appropriate installer (below) for your PC. | |||
 |
|||
[Mac] [Win]
For 64bit version,
If you do not know which version is better, see https://support.microsoft.com/ja-jp/help/958406 (in Japanese) |
|||
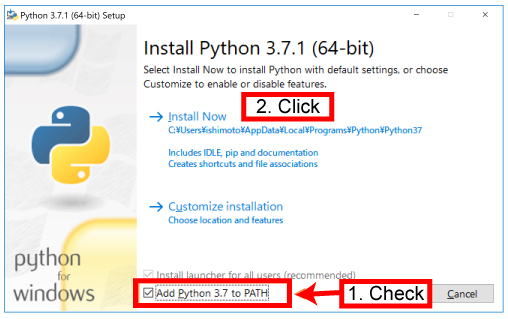
| Then install the package as below. | |||
 |
|||
[Mac]
[Win] |
|||
 |
|||
 |
|||
Alternatively, we can use Command prompt. Find as below:
|
|||
[Mac, Win]
|
|||
Text editors [Mac, Win] |
|||
BBEdit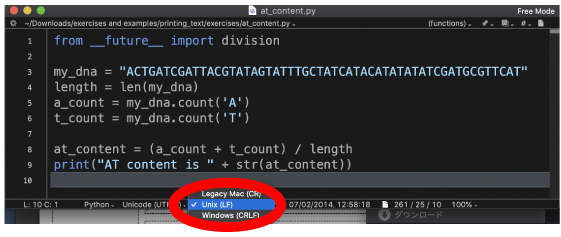 |
|||
Notepad++
|
|||
P14. Hello world We will run hello_world.py from terminal.
We will use the following program:
The underlined part can be obtained by the following step: |
|||
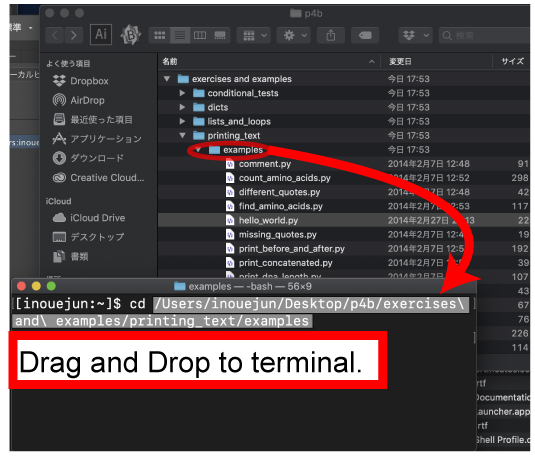
| [Mac, Cygwin] | |||
| Drag and drop the examples icon (shown by red circle) as shown below. | |||
 |
|||
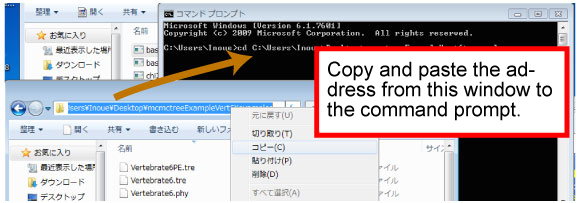
| [Command prompt] | |||
| Copy the address (underlined) as follows. | |||
 |
|||
[Mac, Win]
ls command shows the content of directory:
|
|||
| [Mac, Win] Open hello_world.py by your editor as shown below. |
|||
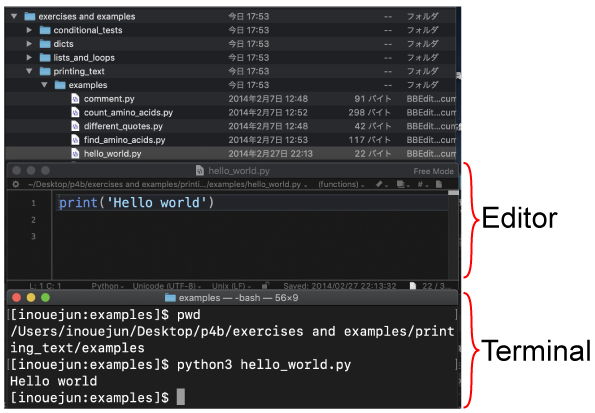 |
|||
Then type,
You will find the following output in your command line.
|
|||
|
|||
P39. Calculating AT content |
|||
 |
|||
At first, we will make sure that the program works.
We will use the following program:
By opening your at_content.py file, make sure the content as below. |
|||
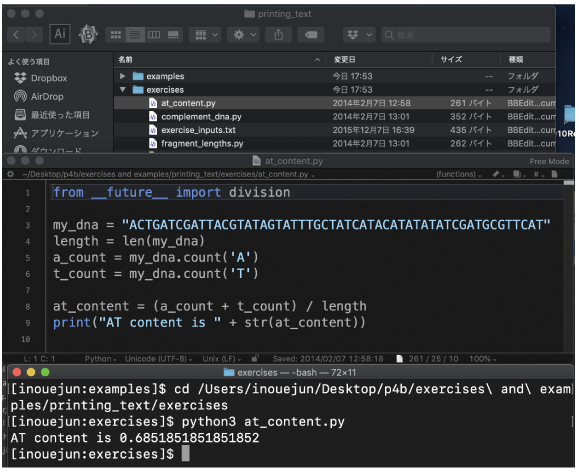 |
|||
Then, run at_content.py:
|
|||
How to study By using your editor (BBEdit/Notepad++), type the following line:
The sequence part, "ACTG..." can be copied from the above or the at_content.py file. Then save this file as test.py in your printing_text/exercises directory. Ignore the first line "from __future__ import division." This is for python2 (p11).
|
|||
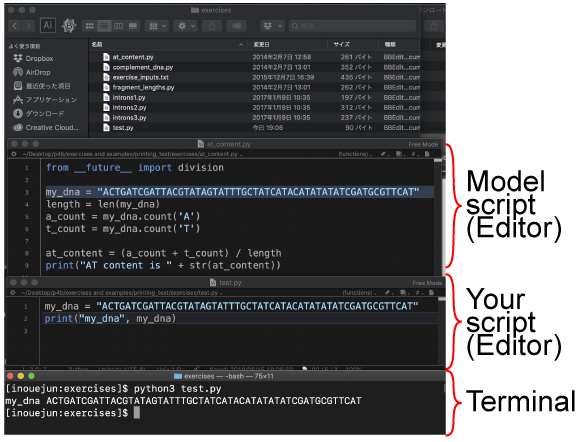 |
|||
For the variable, see P21 "Storing strings in variables."
Again, we will make sure the content of length variable by using the print function. exit() function is useful to make sure variables by stopping programs. |
|||
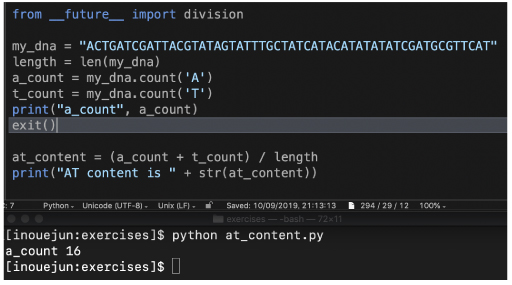 |
|||
P42. Complementing DNA |
|||
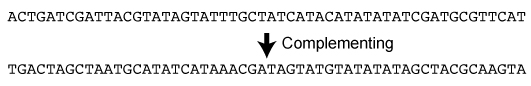 |
|||
cd into the directory:
We will use the following program:
|
|||
|
|||
| P67. Splitting genomic DNA | |||
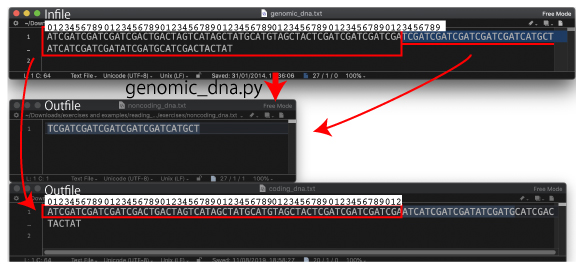 |
|||
Change the directory as follows:
We will use the following program:
|
|||
| P68. Writing a FASTA file | |||
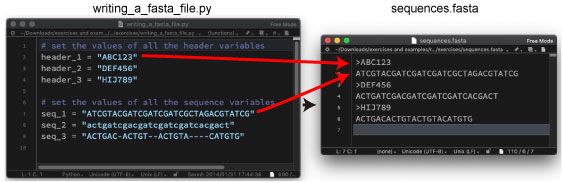 |
|||
We will use the following program:
|
|||
| P72. Writing multiple FASTA file | |||
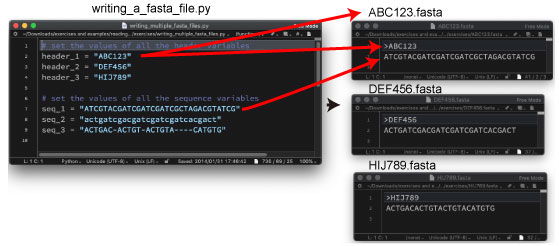 |
|||
We will use the following program:
|
|||
| Application: Reading downloaded fasta file | |||
 |
|||
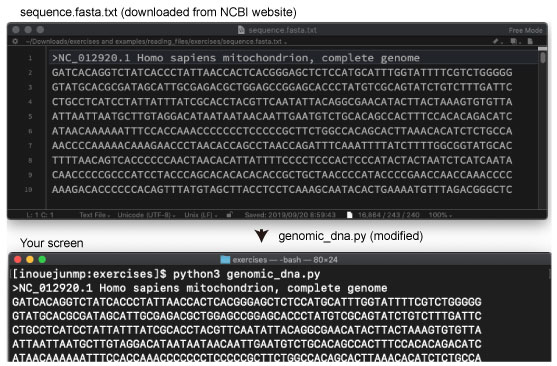
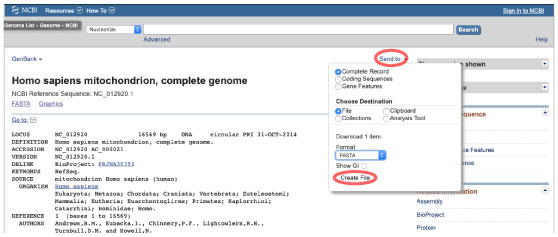
| At first, download human mitochondrial genome sequence from NCBI as below. | |||
 |
|||
| The sequence will be saved as sequence.fasta.txt in your Download directory. Move thie file to the reading_files/exercises directory. Then modify genomic_dna.py to read the sequence.fasta.txt file and print the content in your screen. |
|||
|
|||
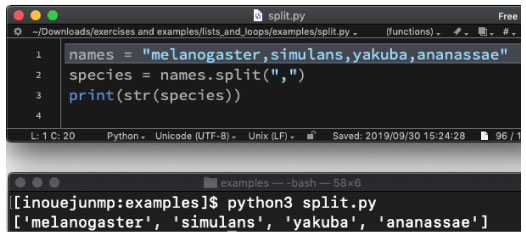
| split.py | |||
 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
| loop.py | |||
 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
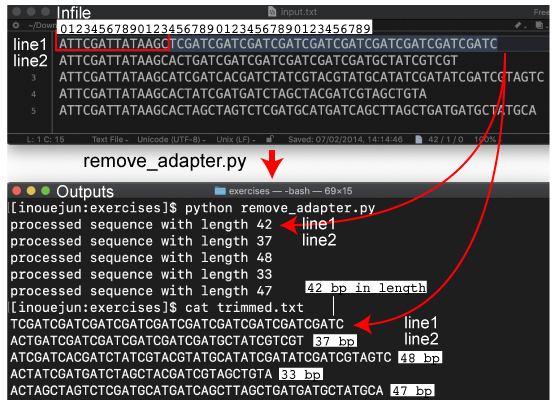
| P90. Processing DNA in a file | |||
 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
| P93. Multiple exons from genomic DNA | |||
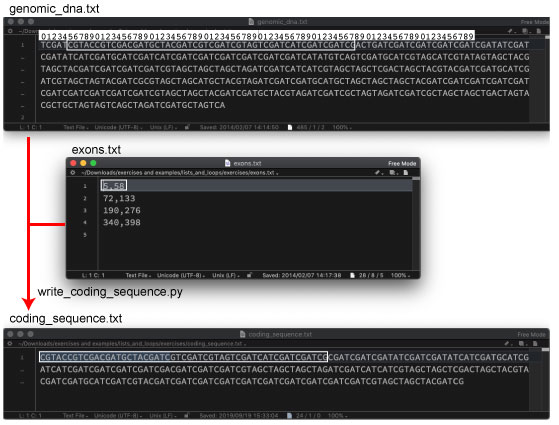 |
|||
We will use the following program:
|
|||
| Loops using excel file outputs | |||
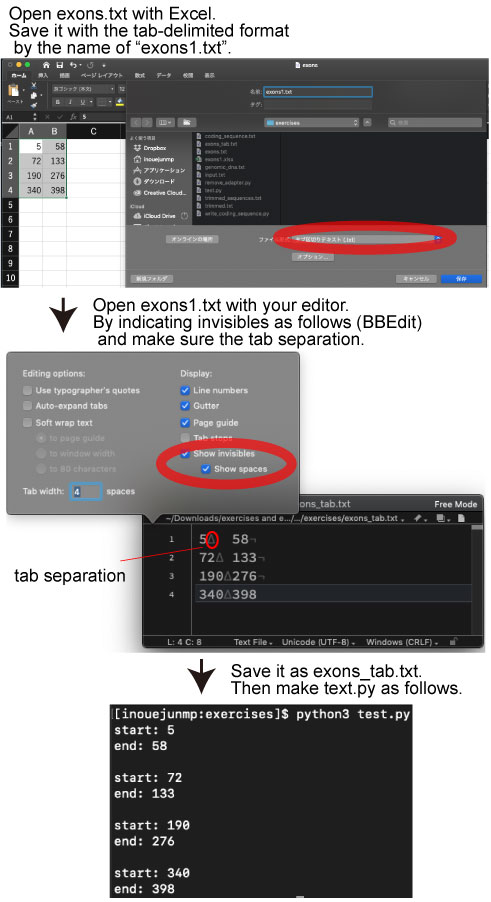 |
|||
We will use the following file:
|
|||
|
|||
| use_function.py | |||
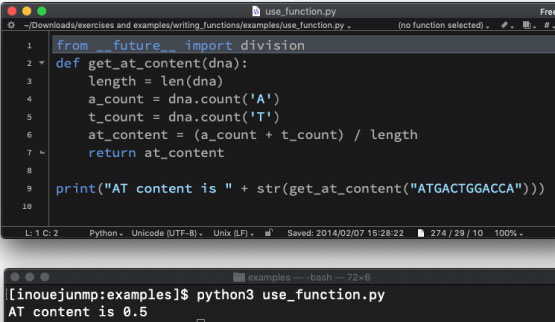 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
| two_arguments.py | |||
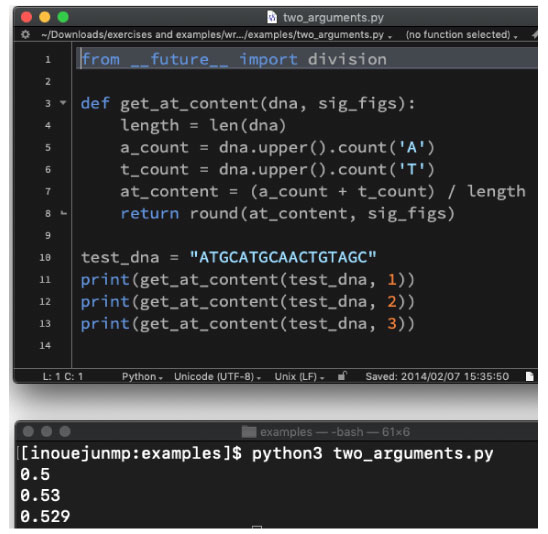 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
| P116. Percentage of amino acid residues, part one | |||
 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
|
|||
| if.py | |||
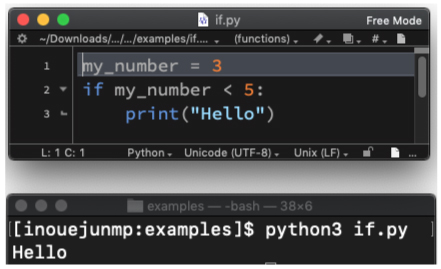 |
|||
cd into the directory by typing the following command:
We will use the following program:
|
|||
| accessions_and.py | |||
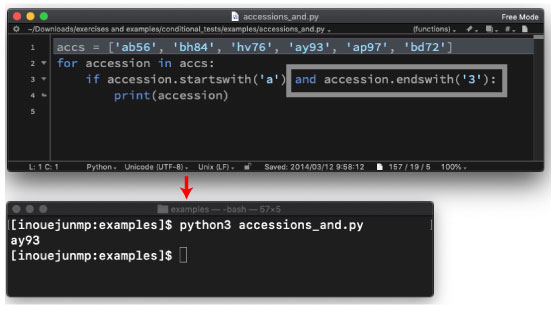 |
|||
We will use the following program:
|
|||
| write_accessions_elif.py | |||
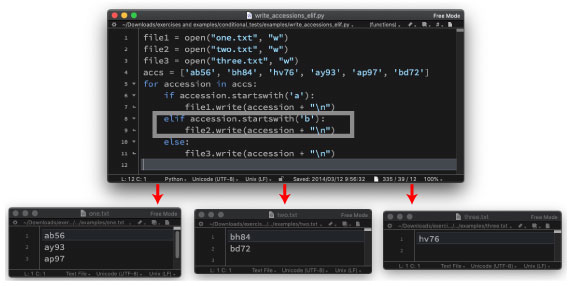 |
|||
We will use the following program:
|
|||
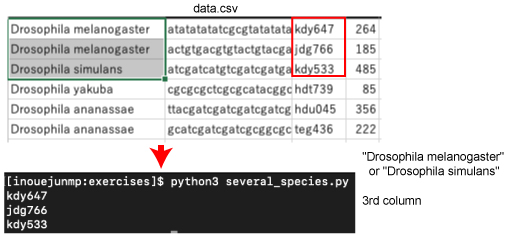
| P135. Several species | |||
cd into the directory by typing the following command:
We will use the following program:
|
|||
| P137. Length range | |||
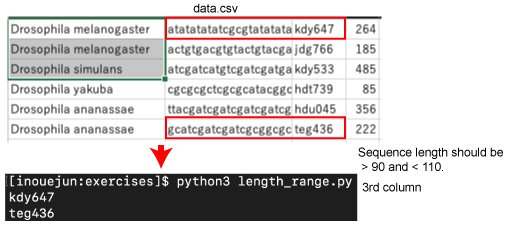 |
|||
We will use the following program:
|
|||
|
|||
List files [Mac]
[Win] dir command.
Changing to the parent directory
Showing full path of the current directory
Opening file with your editor
Scanning images
|
|||
|
|||
|
|