|
BioPerl: Root to tip branch lengths
|
|
|
|
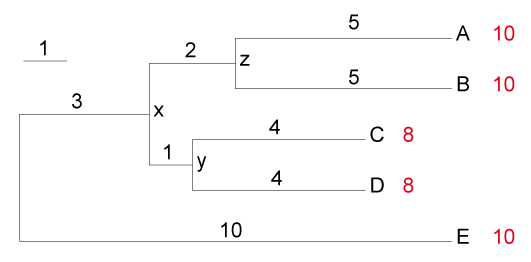
This script calculates the branch lengths between the outgroup and another species. For example, it calculates the branch length between E and A as 20 as below. If the outgroup is not located in the bottom, the user should change the corresponding part of script.
This script was written according to the response from BioPerl mailing list.
|
|
|
|
|
|
#!/usr/bin/perl -w
# Calculate the branche lengths from outgroup to the other species
# perl MyScript.pl
use strict;
use warnings;
use Bio::TreeIO;
use Bio::Tree::TreeFunctionsI;
# read in a phylip/newick format
my $treeio = Bio::TreeIO->new(-format => 'newick', -file => 'treefile.dnd');
my $tree = $treeio->next_tree;
my $rootnode = $tree->get_root_node;
my @leaves = $tree->get_leaf_nodes;
my $og_id = "";
my $og = "";
my $sp_id = "";
my $sp = "";
my $distance = 0;
# Pick up the outgroup.
foreach my $node1 (@leaves) {
$og_id = $node1->id;
$og = $node1;
}
print "Outgroup: $og_id\n";
foreach my $node ( @leaves ) {
$sp_id = $node->id;
$sp = $node;
$distance = $tree->distance( -nodes => [ $og, $sp ] );
print "Distance between $og_id and $sp_id: ";
print "$distance\n";
}
exit;
|
|
|
(((A:5,B:5)z:2,(C:4,D:4)y:1)x:3,E:10);
|
|
|
[inouejun:RootToTip2]$ perl MyScript.pl
Outgroup: E
Distance between E and A: 20
Distance between E and B: 20
Distance between E and C: 18
Distance between E and D: 18
Distance between E and E: 0
|
|
|
|
Dr. Mark A. Jensen gave me suggestions.
|
|
|
Click here.
|
|
|
Dr. Mark A. Jensen's response
My question
Bio::Tree NodeI
[Bioperl-l] suggestions for additions to Tree
|
|